Signatera™ Indications
Signatera™ can be used for a variety of cancer types. To learn more about how, please select your role and a cancer type.
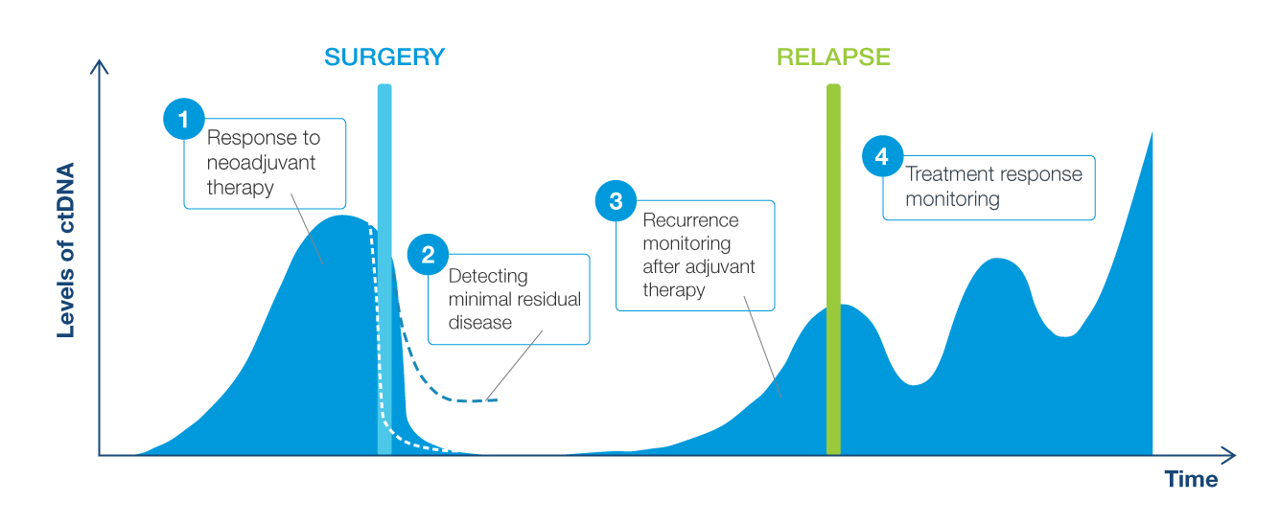
Why circulating tumor DNA (ctDNA) for molecular residual disease assay (MRD) assessment?
- ctDNA is a powerful tool that can be measured to assess the absence or presence of MRD
- Signatera™ is a highly sensitive and personalized MRD assay using ctDNA and is custom designed for each patient to help identify relapse earlier than standard of care tools
Clinical applications of ctDNA testing for MRD assessment

peer-reviewed publications
peer-reviewed publications
Not all MRD tests are created equal
Signatera™ is the most comprehensive MRD test available. Review our peer-reviewed publications here.
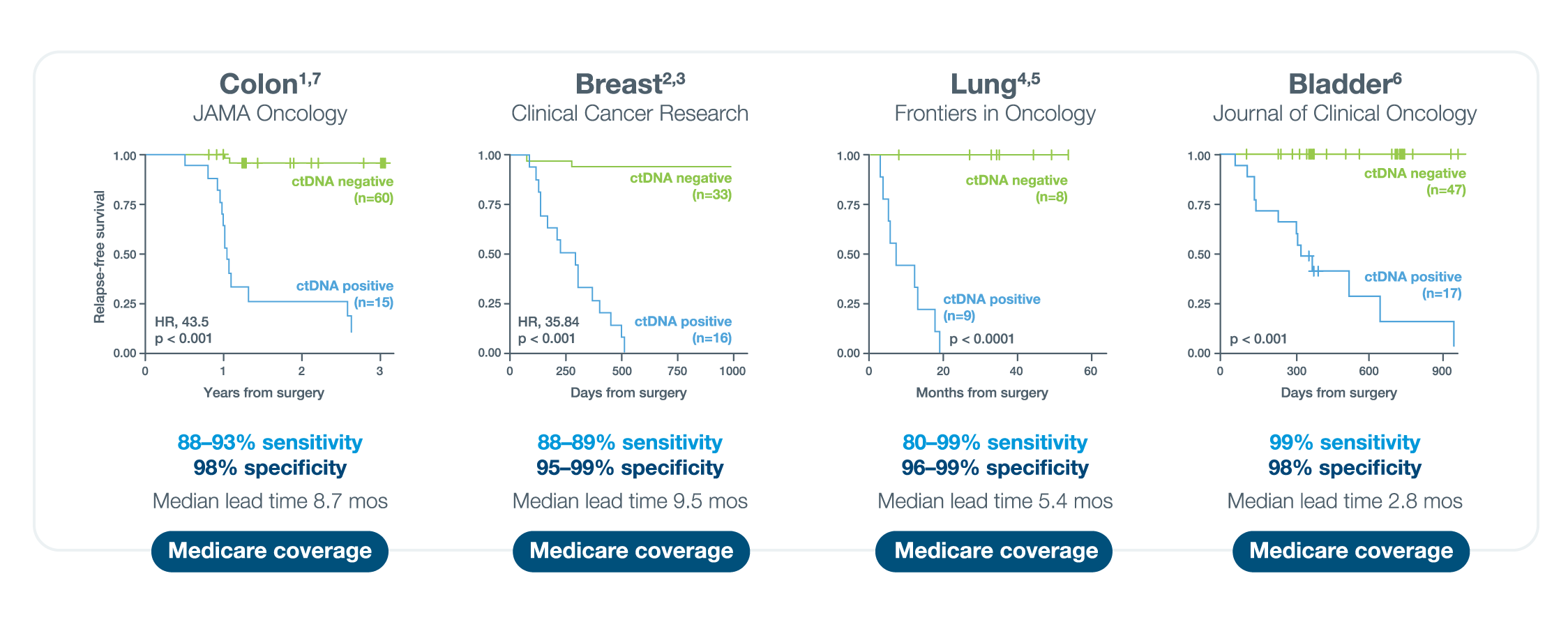
A positive Signatera™ result predicts relapse with overall positive predictive value more than 98%1-4
In clinical studies, Signatera showed high performance across multiple solid tumors
Signatera™ is the first tumor-specific assay for truly individualized cancer care
Personalized design for every patient
- Custom-built assay—based on the unique mutation signature of each patient’s tumor—identifies and tracks tumor mutations at the source
- Once a personalized assay is designed, a patient’s blood can be used to accurately monitor for the presence or absence of the disease over time
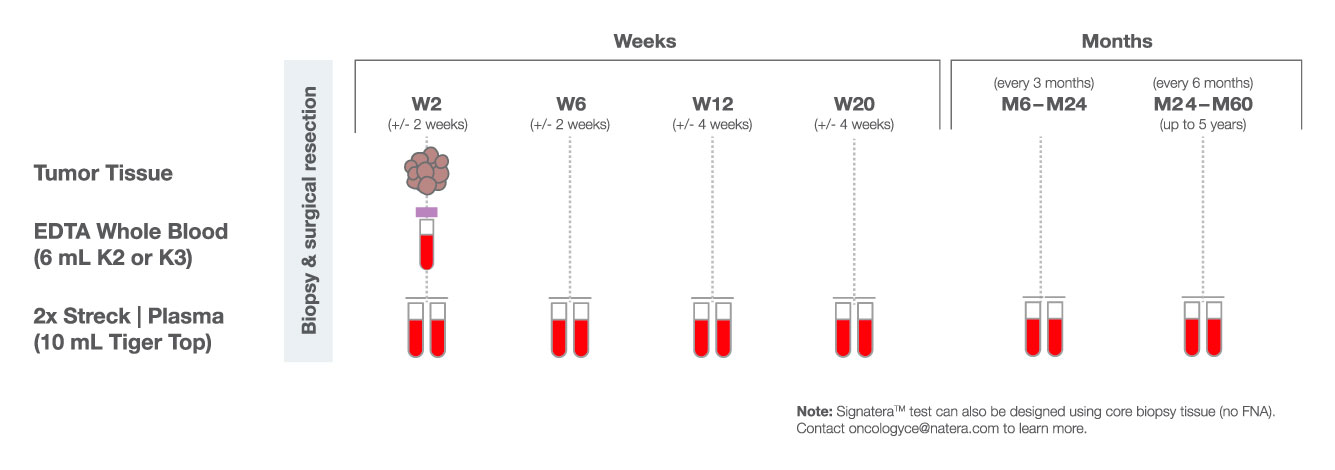
How Signatera™ Works: a personalized and tumor informed approach to MRD surveillance

Personalized, tumor-informed assay
One-time, primary tissue sample and matched normal sample is required for whole exome or whole genome sequencing and personalized test design.
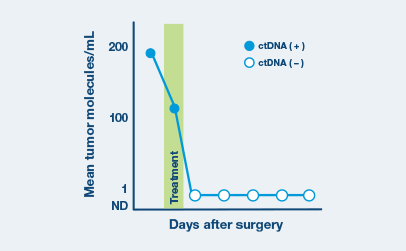
Ultrasensitive ctDNA detection
Signatera™ is designed to detect ctDNA of somatic and truncal variants to optimize sensitivity. Tumor-informed method enables filtering of CHIP mutations to decrease false positive rates.
Optimized for longitudinal monitoring
Once the patient’s personalized test has been designed, only a blood sample is needed each subsequent time.
Recommended ordering cadence of every three months depending on tumor type and clinicians’ preference
To get started, send in your Signatera™ requisition form, sign in to the online portal or talk to your sales representative.
Step 1
Send tumor block or slides to prepare the personalized tumor-informed assay

Step 2
Complete information on tube labels
Step 3
Place filled and labeled tubes into the absorbent sleeve and into the metallic envelope. Place the metallic envelope into the biohazard bag.
Covered by Medicare for multiple solid tumor indications
Is Signatera™ right for you?
We’re here to help you find out
References
1Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stage I to III Colorectal Cancer. JAMA Oncology. 2019;5(8):1124-1131.
2Coombes C, Page K, Salari R, et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clinical Cancer Research. 2019;25(14):4255-4263.
3Abbosh C, Birkbak N, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017,545:446–451
4Christensen E, Birkenkamp-Demtröder K, Sethi H, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients with Urothelial Bladder Carcinoma. 2019; 37(18):1547-1557.






