Assess Disease at the Molecular Level
Signatera™ is a personalized, tumor-informed circulating tumor DNA (ctDNA) assay to detect and monitor molecular residual disease (MRD).

Whole Exome Sequencing
Sequence tumor tissue and matched normal blood to identify tumor-specific genomic signatures
Personalized Assay Design
Create custom multiplex PCR assay to target patient-specific clonal mutations in plasma
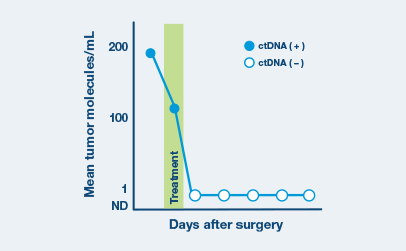
Longitudinal Monitoring
Assess ctDNA levels over time to evaluate disease persistence, recurrence, and treatment response
Enrich for High Risk Populations
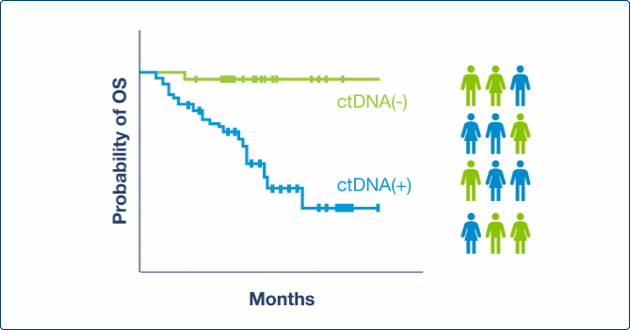
- Stratify & enrich for study participants most likely to benefit from additional treatment1-2
- Treat on molecular relapse at time of lower disease burden
Benefits
- Maximize therapy efficacy and decrease necessary study size
- Move ahead of next-line therapies
Predict Response Sooner
- Predict therapy response as early as 6 weeks3 into treatment
- Support early therapy efficacy readouts as a co-primary or surrogate endpoint
Benefits
- Improve program prioritization and planning
- Accelerate approval of new therapies
… ctDNA as a biomarker has a number of potential regulatory and clinical uses in the early stage setting that may assist and expedite drug development … to enrich a high- or low-risk population for study in a trial, to reflect a patient’s response to treatment, or potentially as an early marker of efficacy …”
FDA DRAFT GUIDANCE FOR INDUSTRY | USE OF CTDNA FOR EARLY-STAGE SOLID TUMOR DRUG DEVELOPMENT MAY 2022
Webinar: Latest data on ctDNA as a patient selection and early efficacy marker for gynecologic cancers
Date: Wednesday, May 29th, 2024
Our expert speakers will discuss the latest data from recent publications and SGO 2024 supporting the use of ctDNA in gynecologic cancer clinical practice and trials.
This will be followed by a panel discussion on the future directions and implications for drug development and clinical trial design in this rapidly evolving space.

Learn About Signatera™ Data for Each Use Case
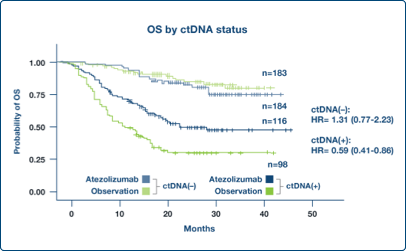
Enrich Populations
Post-surgical ctDNA status identified study participants who benefited from an investigational adjuvant therapy1
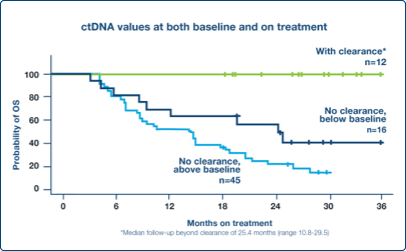
Monitor Response
Early decrease in ctDNA levels at week 6 correlated with favorable outcomes in the metastatic setting3
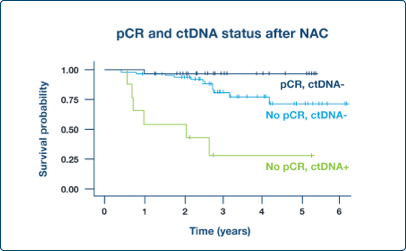
Supplement Endpoints
ctDNA dynamics in neoadjuvant setting can potentially support pathologic complete response (pCR) as surrogate endpoint4
Hear From Experts: Real World Uses
How can Signatera™ support your clinical trials?
Contact us to learn more
References
1 Powles T, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432-437. https://doi.org/10.1038/s41586-021-03642-9
2Oki E, Shirasu H, Watanabe J, et al. Dynamics of circulating tumor DNA after resection of colorectal cancer: Galaxy study in Circulate-Japan. Oral presentation presented at SSO, Dallas, Texas, March 9-12, 2022
3 Bratman SV, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020.1:873-881. https://doi.org/10.1038/s43018-020-0096-5
4Magbanua MJM, et al. Circulating tumor DNA in neoadjuvant treated breast cancer reflects response and survival. Ann Oncol. 2021;32(2):229-239. https://doi.org/10.1016/j.annonc.2020.11.007
5Shoushtari, A, Collins, L, Arranz, E, et al. Early reduction in ctDNA regardless of best RECIST response, is associated with overall survival (OS) on tebentafusp in previously treated metastatic uveal melanoma (mUM) patients. Oral Presentation presented at ESMO GI; Sep 16-21, 2021. Abstract ID: 3600.
6 Christensen E, Birkenkamp-Demtroder K, Sethi H, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol. 2019;37(18):1547-1557.
