Detect and Monitor ctDNA to Inform Treatment Decisions
Identify
High Risk Patients
Signatera™ Genome identified patients
with a
36x
higher risk of recurrence1
Predict Benefit of
Adjuvant Treatment
Signatera™ Genome identified patients
with a
34%
increase in DRFS1
Detect
Recurrence Early
Signatera™ Genome detected distant, extracranial recurrence with
94%
sensitivity and 100% specificity1
Why Signatera™ Genome?
Ultra-sensitive detection down to 1 part per million (PPM)¹
Increased lead times to recurrence
Enhanced sensitivity to detect recurrence
Backed by Signatera™ experience in >1M samples*
Pan-cancer Signatera™ Genome data1
Signatera™ Genome was evaluated in a pan-cancer cohort of 392 patients with breast cancer, NSCLC, melanoma, RCC, and CRC across >2,600 plasma samples1
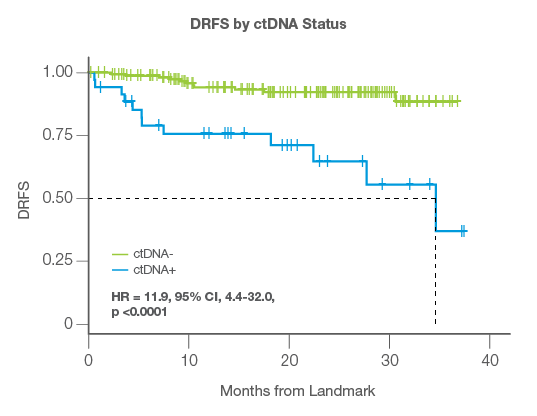
Identify High Risk Patients
- ctDNA status at the landmark time point was highly prognostic of patient outcomes
- ctDNA-positivity post-surgery was associated with significantly worse distant, extracranial relapse-free survival compared to ctDNA-negativity
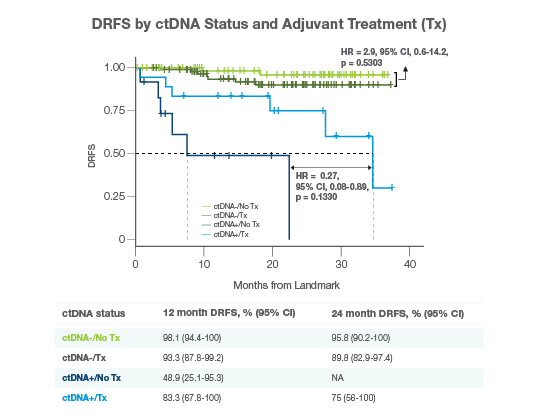
Predict Adjuvant Treatment Benefit
- Patients who were ctDNA-positive after surgery derived a 34% increase in DRFS, while no treatment benefit was observed in ctDNA-negative patients
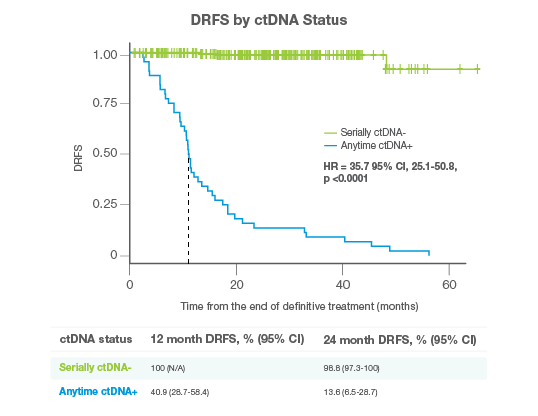
Detect Recurrence Early
- Signatera™ Genome detected distant, extracranial relapse with 94% longitudinal sensitivity and 100% specificity
- ctDNA-positive patients had a 36x higher risk of relapse than ctDNA-negative patients
- Serially ctDNA-negative patients achieved 100% 12-month DRFS vs just 41% for anytime ctDNA-positive patients
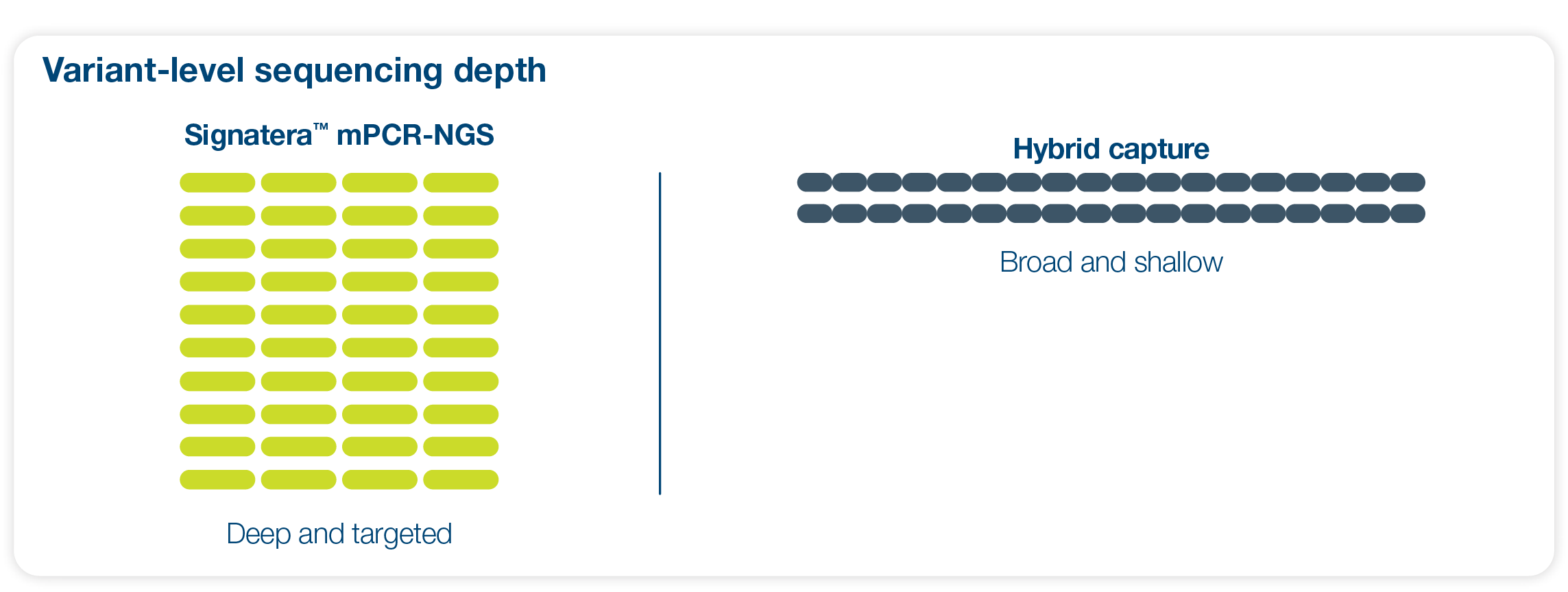
Signatera™’s mPCR-NGS approach
Sequences deeper on the most critical variants, providing the most personalized and clinically meaningful MRD signal
-
Ultra-deep sequencing (up to 350,000x coverage for each selected variant)2
-
Highly curated selection of 64 clonal variants for optimized balance of ultra-sensitivity and high specificity2
-
Proprietary variant selection and calling algorithms for bespoke MRD monitoring2
Natera has the most comprehensive portfolio on MRD solutions to support patient care
Same Signatera™ report; same sample requirements
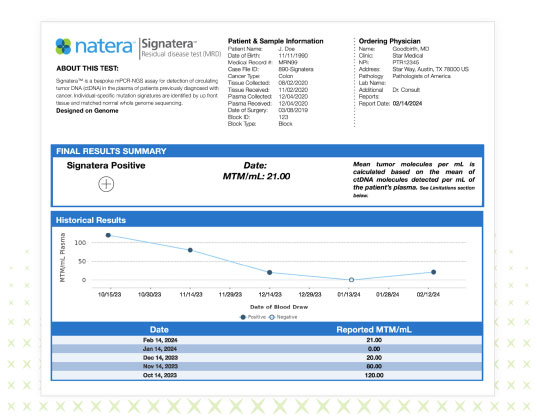
- Patient report: contains the same look and feel as existing Signatera™ report
- Results will be identified as Signatera™ Genome
- TAT: For initial result (4 weeks); 7-10 day TAT for subsequent results
Covered by Medicare for multiple solid tumor indications
Get Started
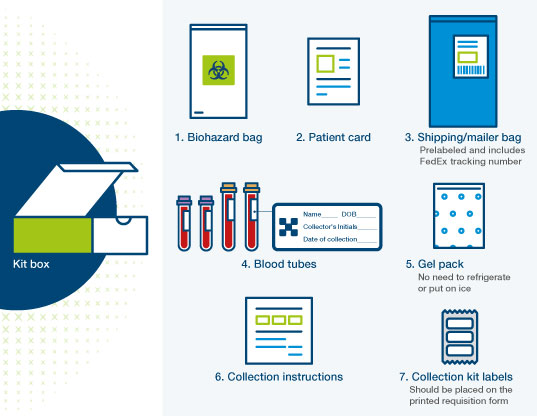
- Signatera™ has the same sample requirements whether designed on genome or exome
- Collect samples as usual and include the printed requisition in the kit
Signatera™ is the most extensively validated and widely adopted MRD assay
Widely Used by Clinicians
Extensive and Robust Clinical Evidence
Is Signatera™ Genome, right for your patients?
We’re here to help you find out
1George M, et al. Poster presented at ASCO Annual Meeting, Chicago, IL, June 2025.
2Natera data on file.